

2024-04-02
On March 29,2024, BRL medicine, in cooperation with the team of Mingyao Liu, Dali Li, and Liren Wang of East China Normal University and the research group of Fanyi Zeng from the School of Basic Medical Sciences of Shanghai Jiao Tong University, published a paper titled "Engineering APOBEC3A deaminase for highly accurate and efficient base editing." The research paper reports the development of three highly accurate cytosine base editors - "haA3A-CBE." This is also another breakthrough in the field of single base editing tool development in less than a year after the BRL medicine team reported new adenine transversion editing tools (AXBEs and ACBEs) in the internationally renowned academic journal Nature Biotechnology in June2023.

Nature Chemical Biology published an article (article online: https://www.nature.com/articles/s41589-024-01595-4)
“haA3A-CBE”: highly efficient and accurate, with extremely low off-target activity
To develop a precise CBE that can achieve efficient base editing in a variety of sequence backgrounds, the research team used hAPOBEC3A to construct A3A-BE4max, rationally designed it based on the crystal structure of hAPOBEC3A, and screened for low off-target through orthogonal R-loop experiments, identifying active mutants.

haA3A-CBE editor structure diagram
Taking into account the on-target editing activity, off-target editing activity, and editing window, the Y130A, VA, and Y130G mutants were selected as high-precision A3A-CBE (haA3A-CBE), named haA3A-CBE-A, haA3A-CBE-VA, and haA3A-CBE-G. To further evaluate the editing activity and window of haA3A-CBE, they were tested in multiple sequence contexts. The results show that the editing activity of haA3A-CBE at methylation sites is significantly higher than that of YE1-BE4max, and the editing efficiency in GC and AC sequences is significantly higher than that of YE1-BE4max and eA3A-BE4max. Their high activity window is located6-7 bits of protospacer. Additionally, the researchers verified that haA3A-CBE has extremely low off-target activity through whole-genome sequencing and RNA sequencing.
Huge potential! “haA3A-CBE” has promising prospects for in vivo gene therapy
To evaluate the efficiency and accuracy of haA3A-CBE in gene therapy, the researchers constructed nine cell lines containing human pathogenic SNVs. After testing, it was found that haA3A-CBE can efficiently and accurately correct6 of the SNVs, showing editing efficiencies of 1.7-4.8 times and 1.9-3.3 times compared to YE1-BE4max on two SNVs located in the GC sequence background. To further test the gene therapy potential of haA3A-CBE, adeno-associated virus (AAV) or lipid nanoparticles (LNP) delivery technology commonly used in gene therapy was employed to treat hereditary tyrosinemia in a mouse model, achieving precise and efficient in vivo editing with a curative effect.
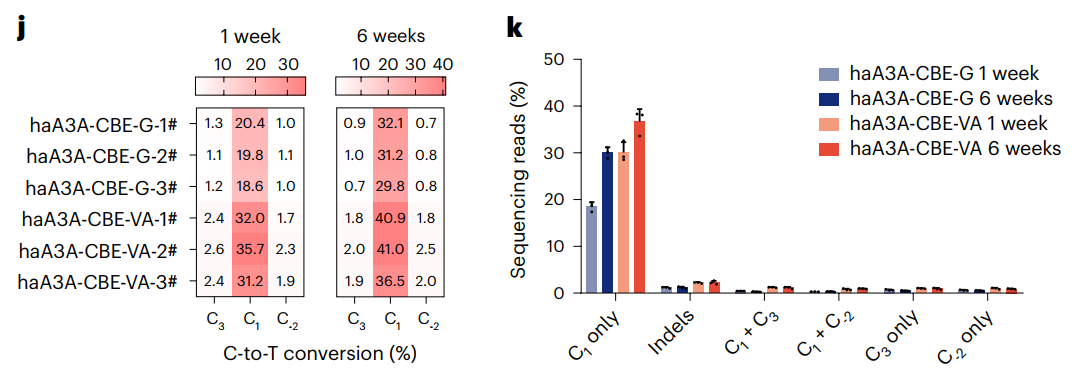
Figure J. Frequency of C to T editing of the FahNS/NS allele per mouse at 1 or 6 weeks after LNP administration. Figure k. FahNS/NS allele editing type and efficiency at 1 or 6 weeks after LNP administration.
Overall, haA3A-CBE has a narrow editing window and extremely low off-target activity. More importantly, it can achieve efficient base editing in a variety of sequence contexts, especially methylation sites and GC sequence background sites. The C base has efficient editing capabilities, enriches the base editing toolbox, and is expected to become a very promising editor for the treatment of human genetic diseases.